A Named Entity Recognition model for molecular dynamics data.
MDNER is a NER model developed specifically to extract information from MD simulations. This project is part of the MDVERSE project [1].
For the GPU code, it's essential to have a relatively new Nvidia GPU that has a minimum memory capacity of 8.0 GiB. It should also be noted that you must have a CUDA driver installed on your system. The code uses CUDA, a parallel computing platform developed by NVIDIA, to interact with GPUs. No specific requirements are needed for the CPU code. To use spaCy, see the spaCy documentation.
Clone the repository and move to the newly created mdner directory:
git clone https://github.com/pierrepo/mdner.git
cd mdner
Install conda and mamba.
Create the mdner and mdner_app conda environments:
mamba env create -f binder/environment.yml
mamba env create -f binder/app.ymlNote: you can also update conda environments with:
mamba env update -f binder/environment.yml
mamba env update -f binder/app.ymlTo deactivate an active conda environment, use:
conda deactivate
This section consists of selecting text datasets (titles and descriptions) that will be used to build our NER model. This consists of generating json files and text files containing the titles and descriptions of our available MD datasets here.
Load the mdner conda environment and launch the generation of text files and json files:
conda activate mdner
python3 scripts/generate_annotation.py
➤ Outputs :
[2023-07-07 17:43:56,748] [INFO] 974 texts selected according the threshold length
[2023-07-07 17:43:59,457] [INFO] 284 texts selected according the corpus similarity
[2023-07-07 17:43:59,545] [INFO] Generation completedThis script does not require a GPU.
usage: generate_annotation.py [-h] [-c] [-p {mbart,bart-paraphrase,pegasus}] [-s SEED] [threshold]
[cutoff]
Generate text and json files in the annotation folder to be used as training sets.
positional arguments:
threshold The threshold for the length of the descriptive texts. By default
the value is 594.
cutoff Select the descriptive texts where the cosine similarity is below
the threshold. By default the value is 0.2.
options:
-h, --help show this help message and exit
-c, --clear Clear the annotation folder and generate files.
-p {mbart,bart-paraphrase,pegasus}, --paraphrase {mbart,bart-paraphrase,pegasus}
Paraphrase the annotation according to three paraphrasing models.
-s SEED, --seed SEED Set the seed for reproducibility in paraphrase. By default the
value is 42.
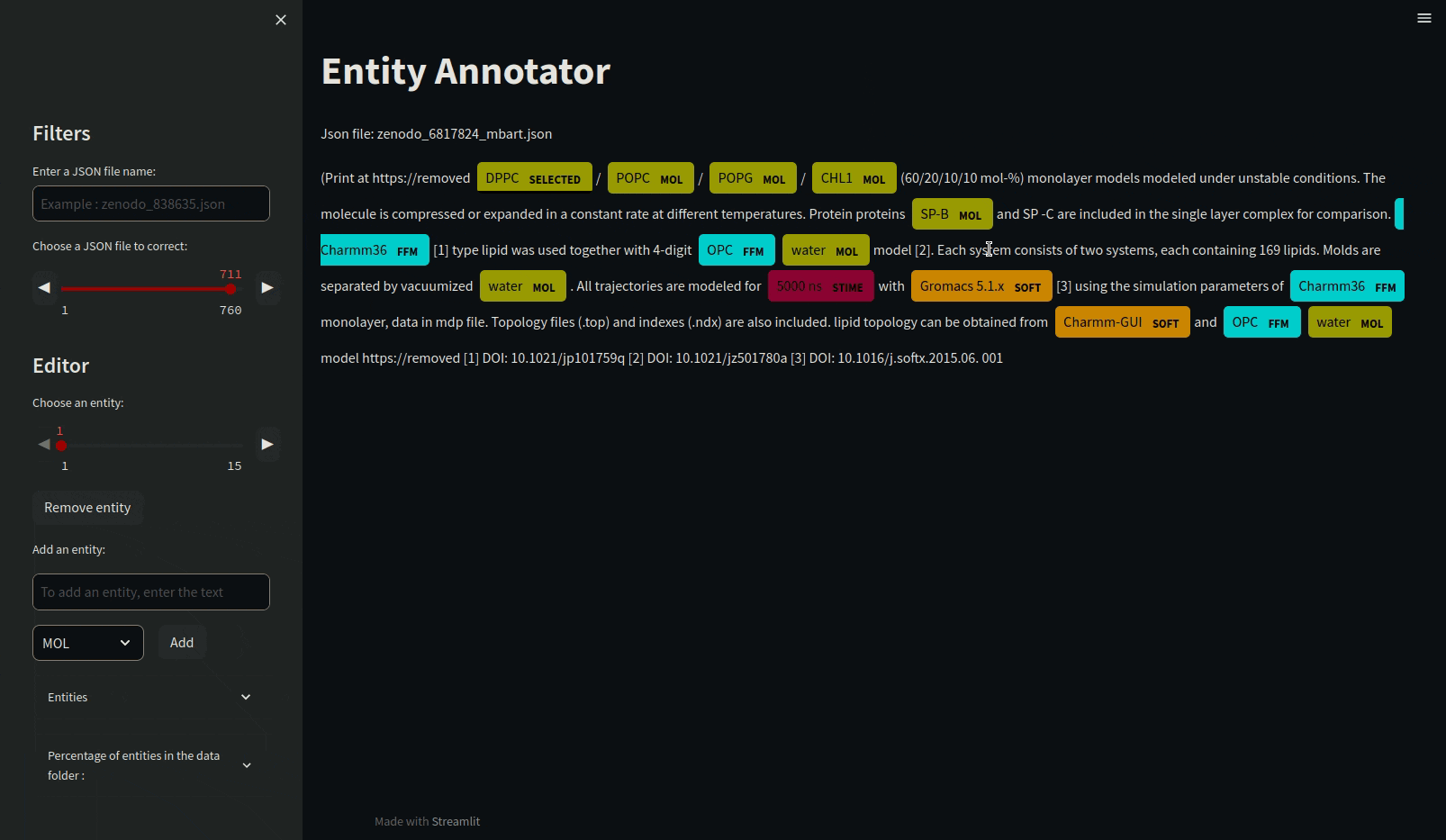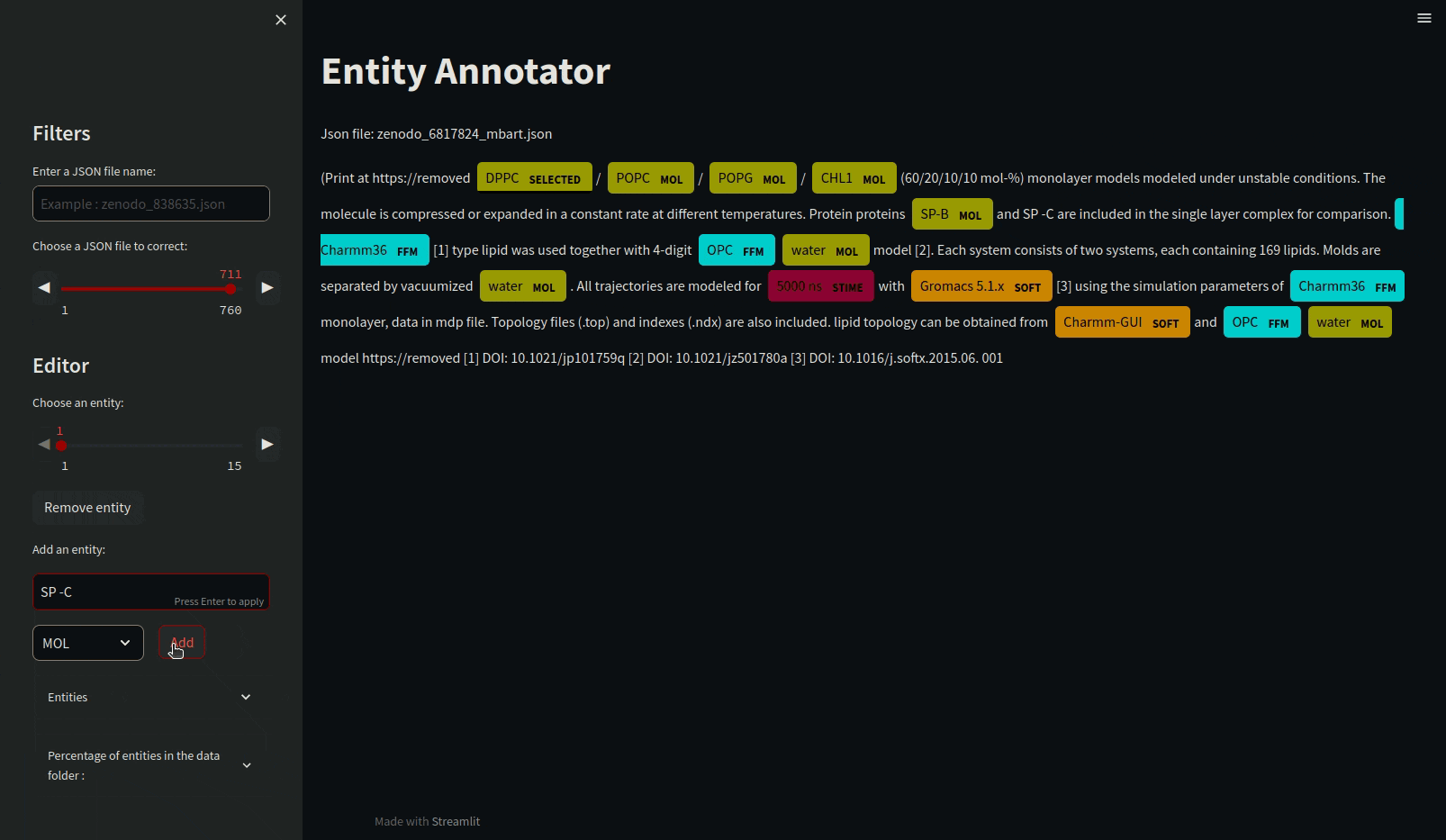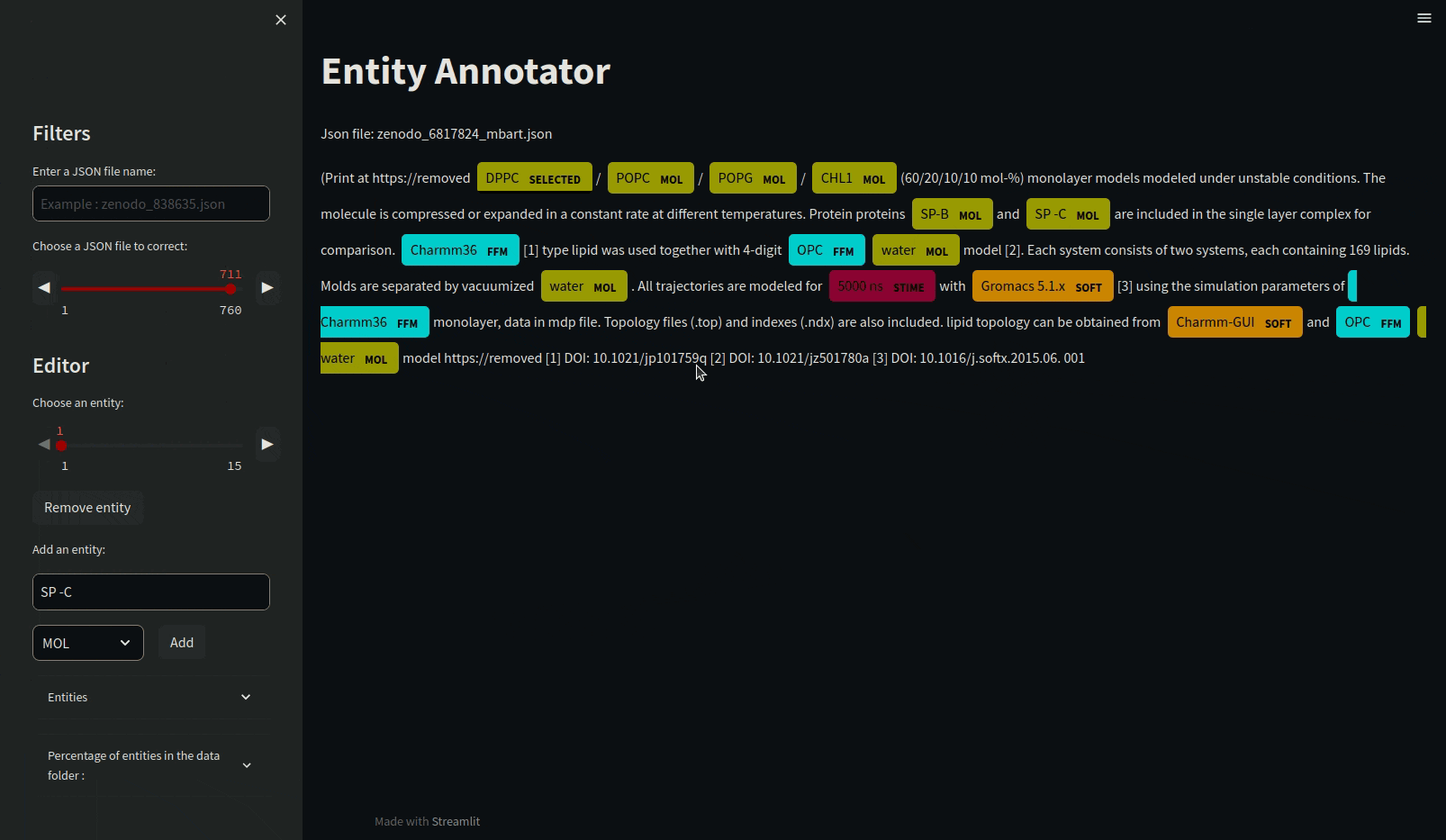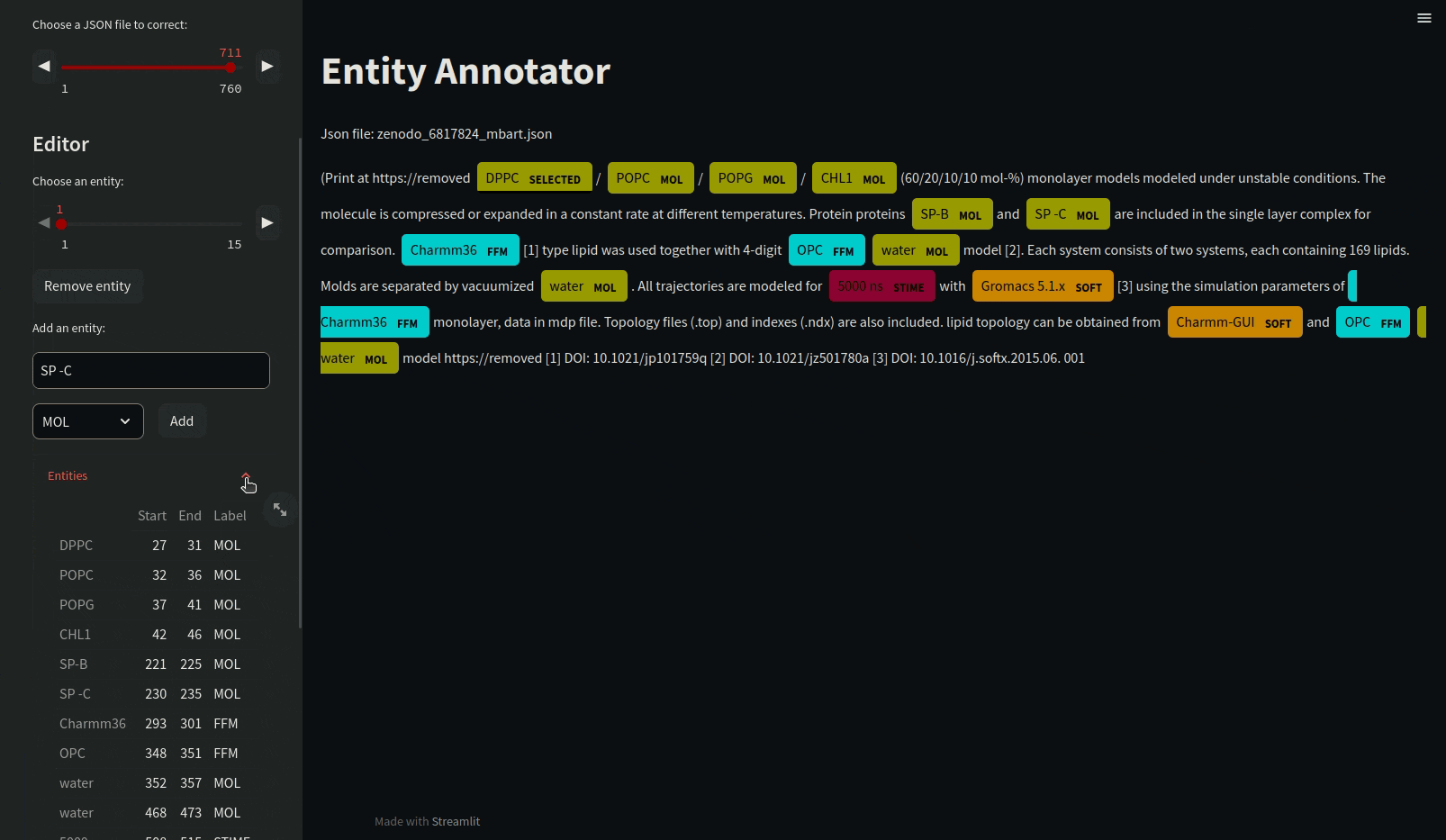
Annotating json files requires manual annotation and must be in the annotations folder. Load the mdner_app conda environment and use the entity annotator to annotate and edit json files by typing the following command :
conda activate mdner_app
streamlit run scripts/entity_annotator.py
There are various other tools for annotating such as Prodigy or a site that allows it: https://tecoholic.github.io/ner-annotator/.
If you think you don't have enough data, you can paraphrase the annotated texts. Paraphrasing consists at keeping the context of the original text and reformulating it in another way.
python3 scripts/generate_annotation.py -p mbartHere you will use the mBART model for paraphrasing.
➤ Output:
[2023-07-21 10:36:56,305] [DEBUG] Starting new HTTPS connection (1): huggingface.co:443
[2023-07-21 10:36:57,272] [DEBUG] https://huggingface.co:443 "HEAD /facebook/mbart-large-50-many-to-many-mmt/resolve/main/config.json HTTP/1.1" 200 0
[2023-07-21 10:37:12,036] [DEBUG] https://huggingface.co:443 "HEAD /facebook/mbart-large-50-many-to-many-mmt/resolve/main/generation_config.json HTTP/1.1" 200 0
[2023-07-21 10:37:12,243] [DEBUG] https://huggingface.co:443 "HEAD /facebook/mbart-large-50-many-to-many-mmt/resolve/main/tokenizer_config.json HTTP/1.1" 200 0
[2023-07-21 10:37:18,072] [INFO] Seed: 42
[2023-07-21 10:37:18,074] [INFO] Paraphrase processing with mbart model: 100%| Files found: 380The execution time for paraphrasing depends on the model used. For mBART, this takes about 2.5 hours with the use of GPU. Paraphrasing could run without GPU, on CPU only, but it will take more time.
A presentation of the annotation structure can be found on ANNOTATIONS.
The mdner.py script is used to create the model according to the defined parameters. Using this script requires a GPU. In our test case, we used an NVIDIA GeForce RTX 3060 12 GB vram.
usage: mdner.py [-h] [-c] [-t d f p r] [-n NAME] [-g] [-p] [-m] [-s SEED]
Create a model for the molecular dynamics data.
options:
-h, --help show this help message and exit
-c, --create Create a dedicated Named Entity Recognition model for our molecular
dynamics data.
-t d f p r, --train d f p r
Hyperparameters for the learning process where d is the percentage
of dropout. The f, p and r scores define what SpaCy believes to be
the best model after the learning process. Each value must be between
0 and 1, and the sum of these three values must be equal to 1.
-n NAME, --name NAME Name of the model.
-g, --gpu Use GPU for learning. Using this parameter, the model that will be
created will be based on the Transfromers "BioMed-RoBERTa-base".
-p, --paraphrase Add paraphrase in the learning dataset.
-m, --mol Use only MOL entities.
-s SEED, --seed SEED Seed used to sample data sets for reproducibility. By default the
value is 42.
To create the mdner, the -c, -t and -n options must be used. The -c option tells the script to create a model. The -t option is the hyperparameters to be used to train the model. The -n option corresponds to the name model that will be created.
You can introduce paraphrases only in the learning set (training + test) with the -p option. Creating a model with a GPU takes more than 1 hour on average, whereas with a CPU it takes around 30 minutes.
conda activate mdner
python3 scripts/mdner.py -c -t 0.1 0.0 1.0 0.0 -n my_model -g -p -s 7522Here, we define a model where the dropout value will be 0.1`` (10% of the nodes will be deactivated). The three next values (0.0 1.0 0.0`) correspond to the metrics f-score, precision and recall. They allow us to consider what is the best model. Here we prefer the precision score rather overt the recall and f- score. The sum of these 3 values must be equal to 1.0.
We have also chosen to create a model based on Transformers by using the -g option. If the -g option is not chosen, the model generated will be based on the cpu and will use a basic spaCy model.
The -p option is used to add paraphrases to the learning dataset.
The -s option specifies the seed used to sample the data sets. You should be able to obtain similar results wit the same seed.
At the end of the code execution, the best NER model will be evaluated on the validation set. The model will be located in the results/models directory. In this example, the model will be in results/models/my_model.
➤ Output:
[2023-07-21 19:43:34,641] [INFO] Seed: 7522
[2023-07-21 19:43:34,645] [INFO] Add paraphrase in the learning dataset
[2023-07-21 19:43:34,682] [WARNING] 42 files ignored because there are not many entities
[2023-07-21 19:43:35,058] [INFO] train_data: 100%| Size: 488
[2023-07-21 19:43:36,442] [INFO] test_data: 100%| Size: 122
[2023-07-21 19:43:36,980] [INFO] eval_data: 100%| Size: 34
[2023-07-21 19:43:37,147] [INFO] Checking GPU availability
[2023-07-21 19:43:38,710] [INFO] GPU is available
[...]
================================== Results ==================================
TOK 100.00
NER P 90.60
NER R 72.29
NER F 80.42
SPEED 3078
=============================== NER (per type) ===============================
P R F
MOL 93.49 70.11 80.13
FFM 88.89 68.09 77.11
SOFT 90.48 80.85 85.39
STIME 72.97 93.10 81.82
TEMP 85.71 66.67 75.00Here, the training phase took about 2 hours with the use of GPU.
From the original and paraphrased texts obtained with the mBART model, we have trained two NER model based on the Transformers "BioMed-RoBERTa-base" and we evaluated the models on the validation set as shown in Table 1. The models were obtained on the seed 7522 and 10 replicates were generated for each of the 2 models. These replicates took more than 20 hours to run. The results obtained are available in the file results/outputs/results.csv and the bash script scripts/build is used to create the different models :
bash scripts/build.sh| Entities |
Precision score (%) |
|
|---|---|---|
| Transformers | Transformers + Paraphrase | |
| MOL (molecule) | 80 ± 1.9 | 91 ± 1.3 |
| FFM (force field & model) | 75 ± 12.8 | 94 ± 2.8 |
| TEMP (temperature) | 46 ± 17.6 | 90 ± 0.0 |
| STIME (simulation time) | 62 ± 9.7 | 82 ± 3.6 |
| SOFT (software) | 89 ± 7.5 | 92 ± 3.3 |
| Total | 78 ± 1.9 | 91 ± 1.0 |
We note an increase in the precision score, particularly for our key entity, the MOL entity, which rises from 80% to 91%. Performance for the other entities is improved. The NER models were able to identify molecule names not present in the learning dataset, perfectly underlining the ability of the NER model to generalize and identify the desired entities, and demonstrating the relevance of fine-tuning on Transformer models [2].
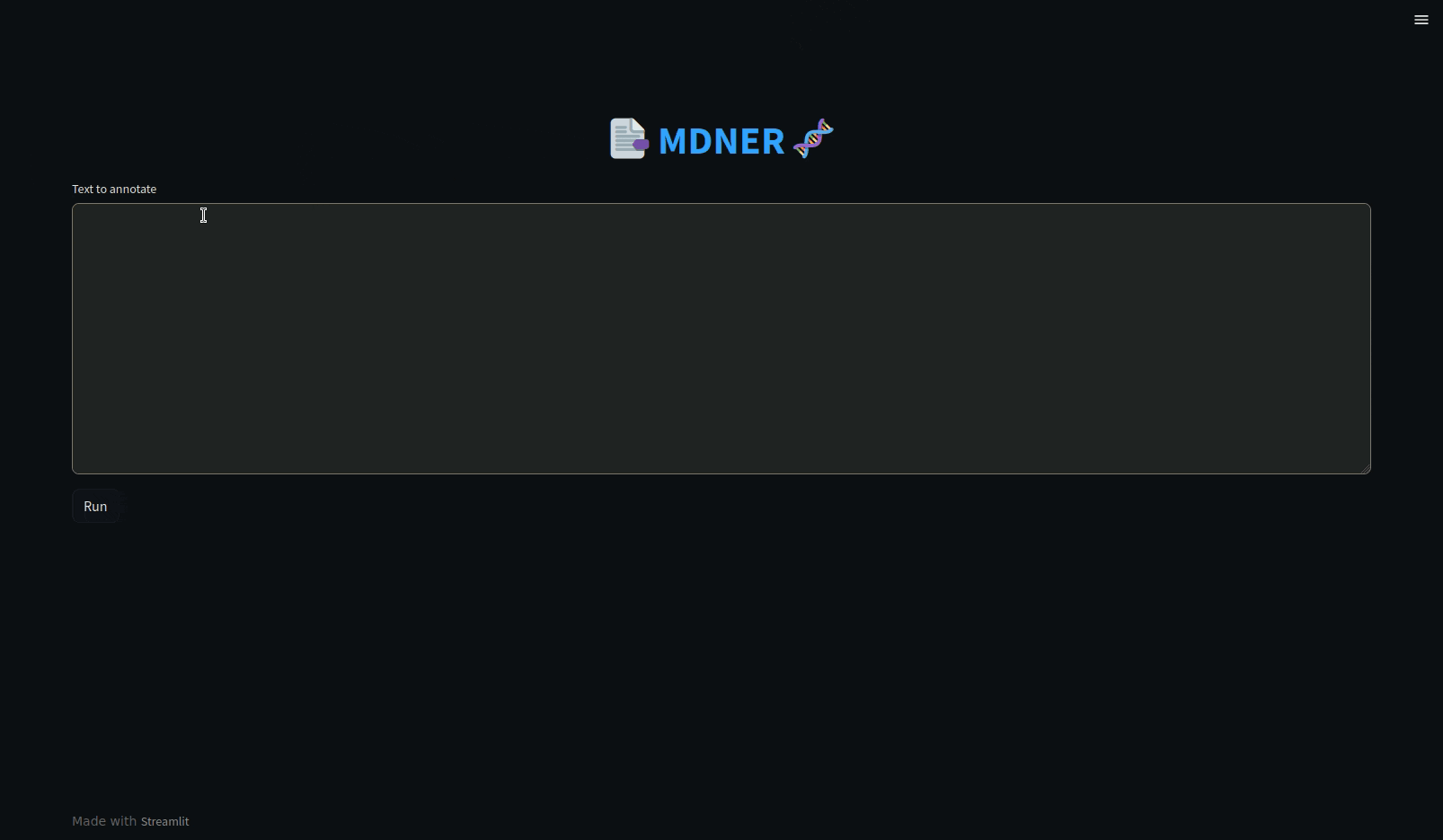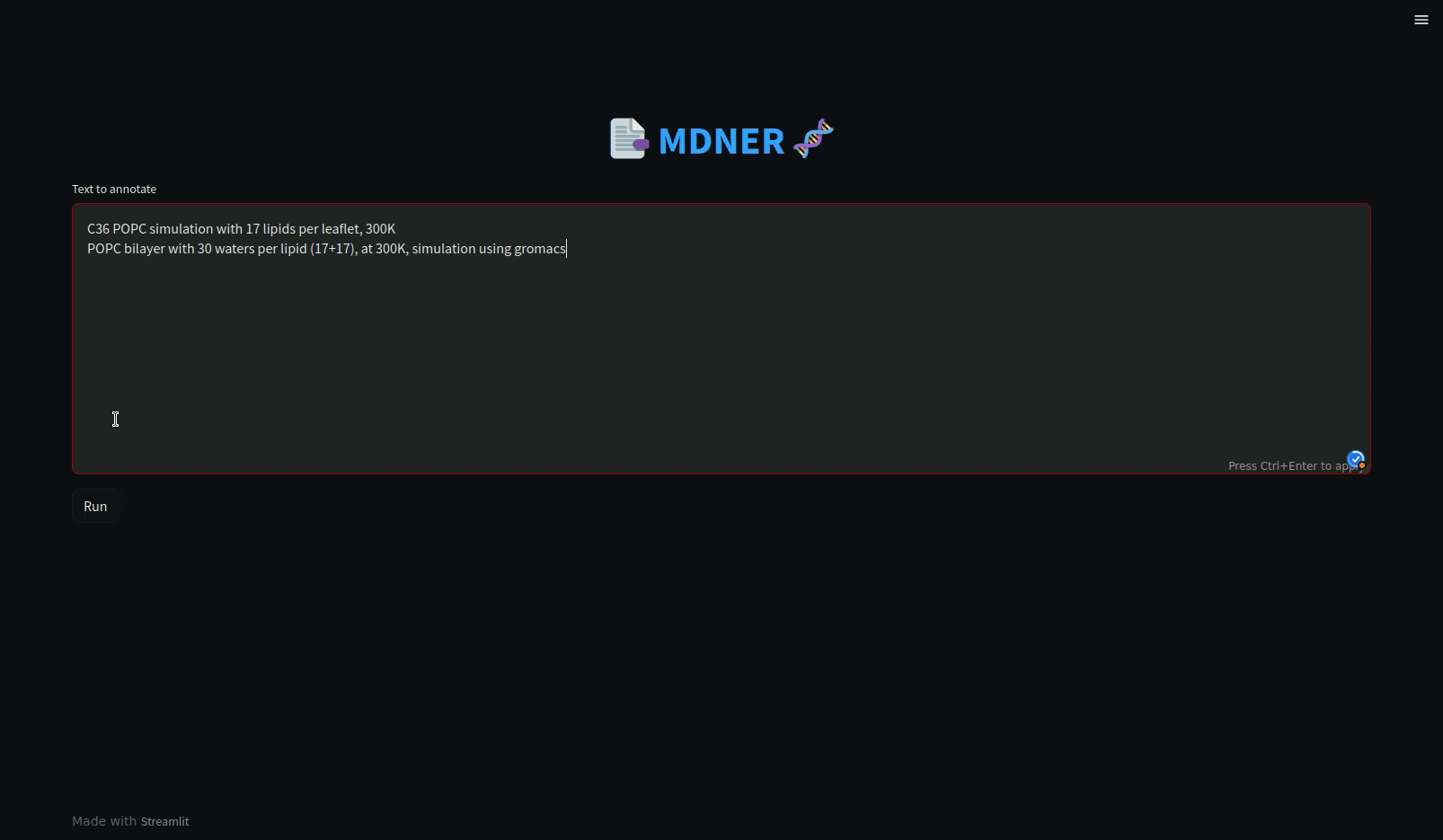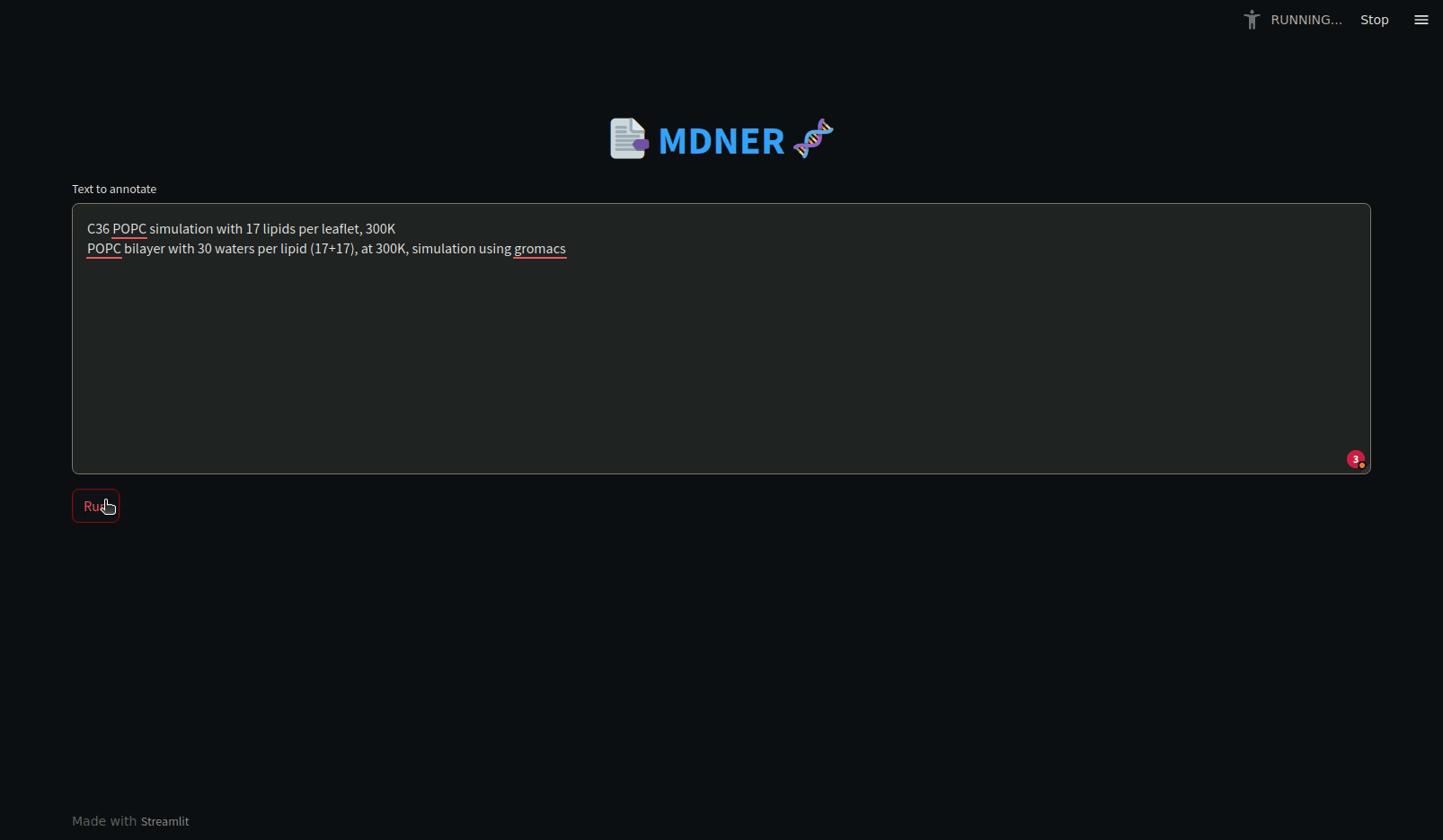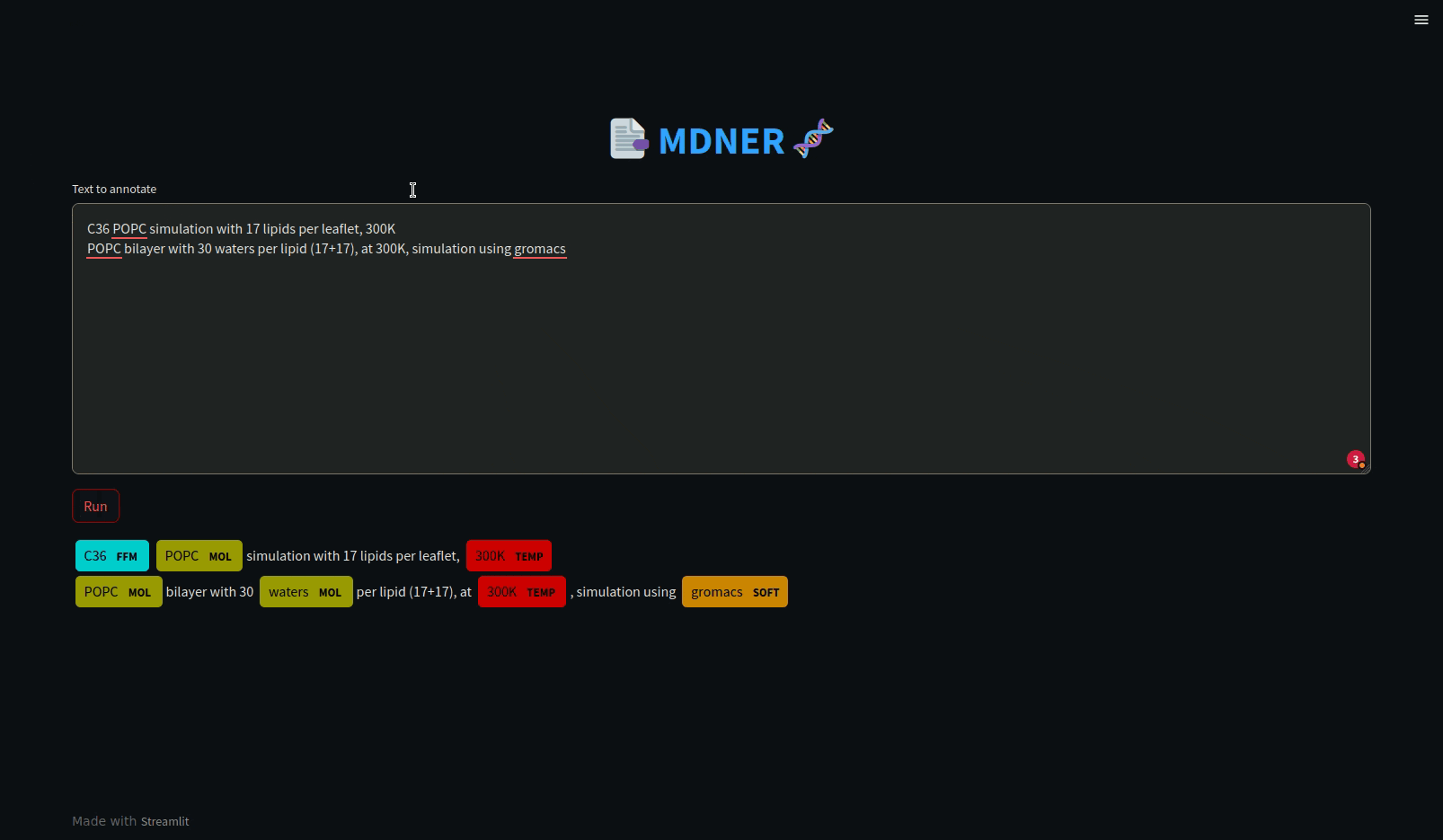
In order to run an example, you can launch a website with Streamlit to apply the MDNER model to a text and annotate it automatically. Load the mdner_app conda environment and run the Streamlit app by entering the name of the model as an argument, as in the following command:
conda activate mdner_app
streamlit run scripts/mdner_app.py -- --model my_model
Using MDNER does not require a GPU. It may be advantageous to use a GPU to speed up the predictions of the NER model, but it is not mandatory.
[1] Tiemann JKS, Szczuka M, Bouarroudj L, Oussaren M, Garcia S, Howard RJ, Delemotte L, Lindahl E, Baaden M, Lindorff-Larsen K, Chavent M, Poulain P. MDverse: Shedding Light on the Dark Matter of Molecular Dynamics Simulations. bioRxiv [Preprint]. 2023 May 2:2023.05.02.538537. doi: 10.1101/2023.05.02.538537. PMID: 37205542; PMCID: PMC10187166.
[2] Suchin Gururangan, Ana Marasović, Swabha Swayamdipta, Kyle Lo, Iz Beltagy, Doug Downey, and Noah A. Smith. 2020. Don’t Stop Pretraining: Adapt Language Models to Domains and Tasks. In Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics, pages 8342–8360, Online. Association for Computational Linguistics.